The SAAVpedia is a comprehensive proteogenomic interpretation platform that identifies, annotates, and retrieves SAAVs from proteomic and genomic sequence data to discover true pathogenic variant candidates. SAAVpedia provides a proteogenomic analysis pipeline to prioritize and interpret SAAVs.
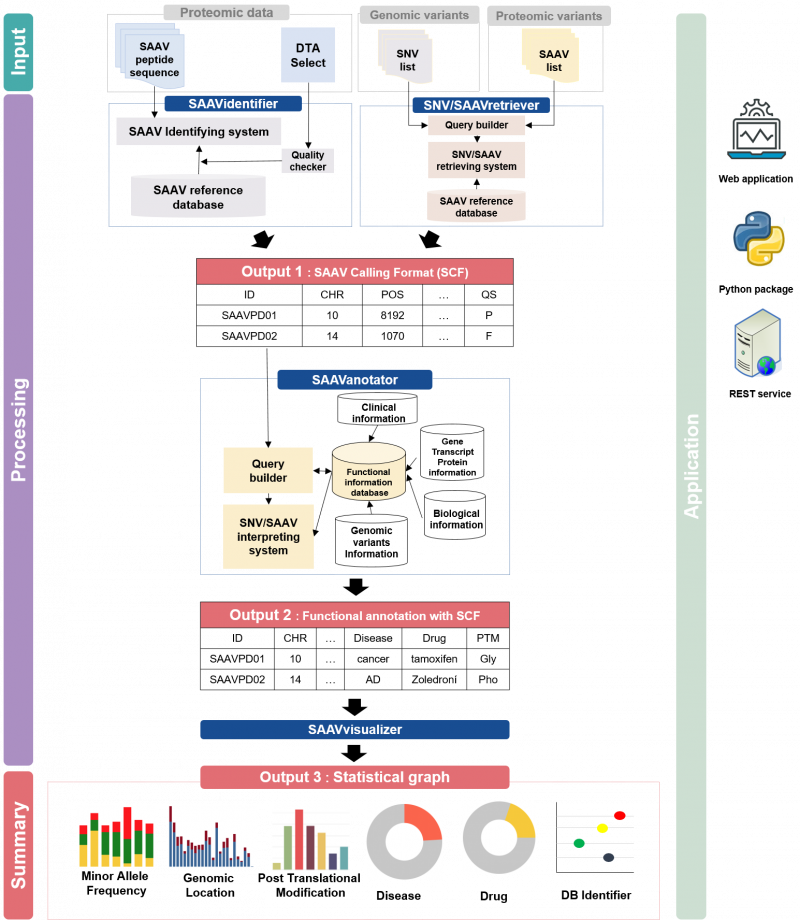
The pipeline contains a database for proteome searches, a SAAV quality checker, a query builder based on clinical, biological, and pharmacological information, and a retrieving and interpreting system. The platform consists of four analysis modules provided by the SAAVpedia web, stand-alone, and REST applications.
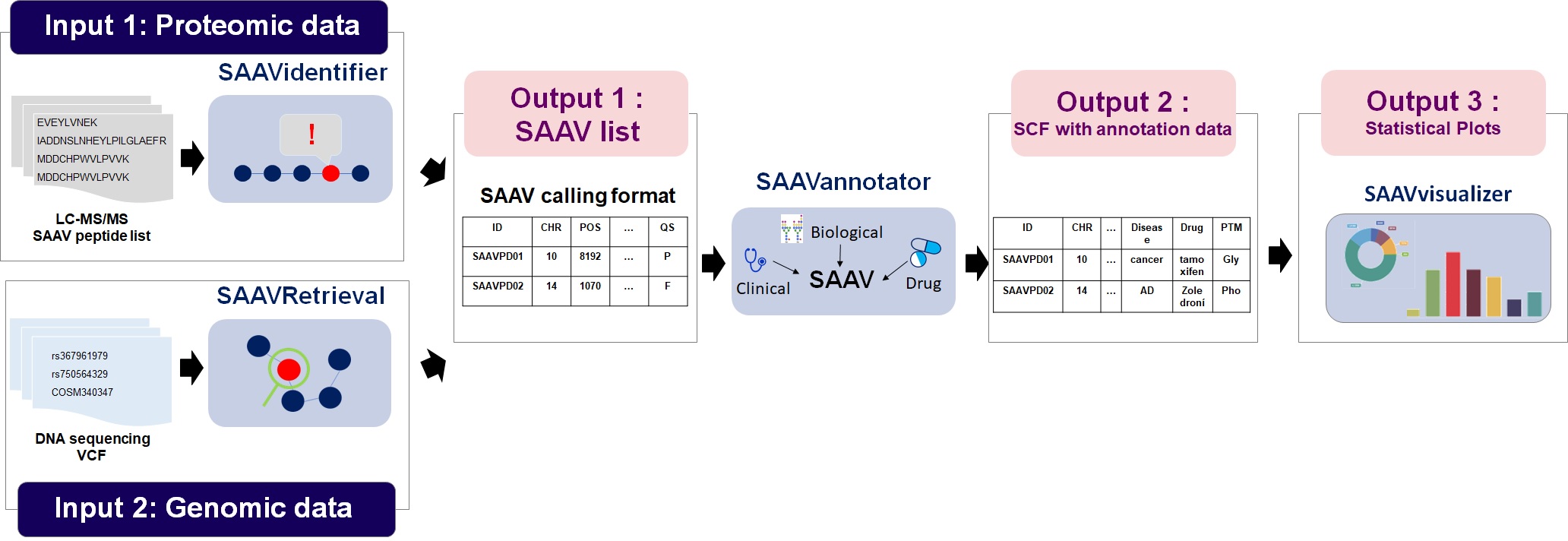
SAAVidentifier module automatically performs simultaneous detection, quality control, and assessment of massive amounts of SAAVs from peptide sequencing data. The results are provided by the SAAV-calling format (SCF) that we designed to facilitate information transmission, by which the text file format contains meta-information including the amino acid sequence and protein and genomic position of each SAAV.
SAAVannotator module performs pharmacological, clinical, and biological annotation based on the genomic and proteomic location of identified SAAVs, e.g., annotating phenotype-variant relationships, post-translational modifications (PTMs), and various kinds of variant or gene database identifiers (IDs) such as dbSNP and Ensembl.
SNVretriever module searches for SAAVs that are in the same location as input nsSNV genomic variants and provides the variant list along with diverse information including diseases and drugs. Otherwise, the SAAVretriever module enables querying and navigating SAAVs that were identified from different proteomic data of various phenotype conditions.
SAAVvisualizer module provides statistical summary plots based on the results of the SAAVannotator and SNV/SAAVretriever modules.
SAAVpedia User World Map (by Google Analytics)
(Jan 1st, 2018 ~ Dec 31th, 2020)
(Jan 1st, 2018 ~ Dec 31th, 2020)
Latest News:
2020.08.31. In the neXtProt website, we now provide information from SAAVpedia for each neXtProt entry. Example: APOE NX_P02649 NEWS on the neXtProt.org Click!! Here
2020.07.24. Collaboration with neXtProt is in progress. – github
2019.12.24. Congratulations! A paper of SAAVpedia is published in J. Proteome Res. click here!
2018.11.28. SAAVpedia User Guide (manual) 2nd version is released.
2018.05.30. SAAVpedia User Guide (manual) first version is released.
2018.01.05. Welcome to SAAVpedia! First release of SAAVpedia.
Browser compatibility (It was tested on November 9th, 2018.)
| OS | Version | Chrome | Firefox | Microsoft Edge | Safari |
|---|---|---|---|---|---|
| Linux | Ubuntu 16.04 | 70.0.3538.77 | 63.0 | n/a | n/a |
| MacOS | Sierra 10.12.6 | 70.0.3538.77 | 63.0 | n/a | 12.0.1 |
| MS Windows | 10 | 70.0.3538.77 | 63.0 | 42.17134.1.0 | n/a |
Acknowledgements :
National Research Council of Science and Technology, Rep. of Korea (CAP-15-03-KRIBB)
The Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health and Welfare, Rep. of Korea (HI13C2098)
Korea Basic Science Institute, Rep. of Korea (T39710)
neXtProt (www.neXtProt.org)